Contents
function results = trial_matrixCompletion(optIn)
setup_MC
if nargin == 0
clc
defaultStream = RandStream.getGlobalStream;
if 1
savedState = defaultStream.State;
save random_state.mat savedState;
else
load random_state.mat
end
defaultStream.State = savedState;
optIn.tryBigampFastApproximated3 = 1;
optIn.tryBigamp = 1;
optIn.tryBigampEM = 1;
optIn.tryBigampEMcontract = 1;
optIn.tryInexactAlm = 1;
optIn.tryGrouse = 1;
optIn.tryLmafit = 1;
optIn.tryVbmc = 1;
optIn.tryMatrixAlps = 1;
optIn.SNR = inf;
optIn.M = 500;
optIn.L = 500;
optIn.N = 10;
optIn.p1 = 0.1;
end
Problem Setup
tryBigamp = optIn.tryBigamp;
tryBigampFastApproximated3 = optIn.tryBigampFastApproximated3;
tryBigampEM = optIn.tryBigampEM;
tryBigampEMcontract = optIn.tryBigampEMcontract;
tryLmafit = optIn.tryLmafit;
tryInexactAlm = optIn.tryInexactAlm;
tryGrouse = optIn.tryGrouse;
tryVbmc = optIn.tryVbmc;
tryMatrixAlps = optIn.tryMatrixAlps;
SNR = optIn.SNR;
M = optIn.M;
L = optIn.L;
N = optIn.N;
p1 = optIn.p1;
checkVal = (M + L).*N ./ (p1 .* M .* L);
disp(['Check condition was ' num2str(checkVal)])
rho = N*(L + M - N) / p1 / M / L;
disp(['Rho was ' num2str(rho)])
Check condition was 0.4
Rho was 0.396
Define options
opt = BiGAMPOpt;
if p1 <= 0.2
opt.sparseMode = 1;
end
Build the true low rank matrix
X = randn(N,L);
A = randn(M,N);
Z = A*X;
Form the (possibly noisy) output channel
error_function = @(qval) 20*log10(norm(qval - Z,'fro') / norm(Z,'fro'));
opt.error_function = error_function;
nuw = norm(reshape(Z,[],1))^2/M/L*10^(-SNR/10);
Y = Z + sqrt(nuw)*randn(size(Z));
omega = false(M,L);
ind = randperm(M*L);
omega(ind(1:ceil(p1*M*L))) = true;
Y(~omega) = 0;
problem = BiGAMPProblem();
problem.M = M;
problem.N = N;
problem.L = L;
[problem.rowLocations,problem.columnLocations] = find(omega);
Establish the channel objects for BiG-AMP
gX = AwgnEstimIn(0, 1);
gA = AwgnEstimIn(0, 1);
if opt.sparseMode
gOut = AwgnEstimOut(reshape(Y(omega),1,[]), nuw);
else
gOut = AwgnEstimOut(Y, nuw);
end
Warning: Tiny non-zero variances will be used for computing log likelihoods. May
cause problems with adaptive step size if used.
Control initialization
Ahat = randn(M,N);
xhat = randn(N,L);
opt.xhat0 = xhat;
opt.Ahat0 = Ahat;
opt.Avar0 = 10*ones(M,N);
opt.xvar0 = 10*ones(N,L);
results = [];
Try BiG-AMP Lite
if tryBigampFastApproximated3
disp('Starting BiG-AMP Lite')
tstart = tic;
[~,~,estHist5] = ...
BiGAMP_Lite(Y,1,1,nuw,problem,opt);
tGAMP5 = toc(tstart);
loc = length(results) + 1;
results{loc}.name = 'BiG-AMP Lite';
results{loc}.err = estHist5.errZ(end);
results{loc}.time = tGAMP5;
results{loc}.errHist = estHist5.errZ;
results{loc}.timeHist = estHist5.timing;
end
Starting BiG-AMP Lite
Try BiGAMP
if tryBigamp
disp('Starting BiG-AMP')
tstart = tic;
[estFin,~,estHist] = ...
BiGAMP(gX, gA, gOut, problem, opt);
tGAMP = toc(tstart);
loc = length(results) + 1;
results{loc}.name = 'BiG-AMP';
results{loc}.err = estHist.errZ(end);
results{loc}.time = tGAMP;
results{loc}.errHist = estHist.errZ;
results{loc}.timeHist = estHist.timing;
end
Starting BiG-AMP
Try EM-BiGAMP
if tryBigampEM
disp('Starting EM-BiG-AMP')
tstart = tic;
[estFin,~,~,estHistEM] = ...
EMBiGAMP_MC(Y,problem,opt);
tGAMP = toc(tstart);
loc = length(results) + 1;
results{loc}.name = 'EM-BiG-AMP';
results{loc}.err = opt.error_function(estFin.Ahat*estFin.xhat);
results{loc}.time = tGAMP;
results{loc}.errHist = estHistEM.errZ;
results{loc}.timeHist = estHistEM.timing;
end
Starting EM-BiG-AMP
It 0001 nuX = 9.906e-01 meanX = 0.000e+00 tol = 1.000e-04 nuw = 9.906e-02 SNR = 20.00 Error = -40.7539 numIt = 0052
It 0002 nuX = 8.911e-01 meanX = 0.000e+00 tol = 2.737e-05 nuw = 2.717e-04 SNR = 45.63 Error = -73.9313 numIt = 0031
It 0003 nuX = 8.941e-01 meanX = 0.000e+00 tol = 1.000e-08 nuw = 7.555e-08 SNR = 81.22 Error = -141.0238 numIt = 0055
It 0004 nuX = 8.940e-01 meanX = 0.000e+00 tol = 1.000e-08 nuw = 3.268e-08 SNR = 149.18 Error = -152.8152 numIt = 0030
Try EM-BiGAMP with rank contraction
if tryBigampEMcontract
EMopt.learnRank = 2;
opt.nit = 300;
disp('Starting EM-BiG-AMP with rank contraction')
tstart = tic;
[estFin,~,~,estHistEM] = ...
EMBiGAMP_MC(Y,problem,opt,EMopt);
tGAMP = toc(tstart);
loc = length(results) + 1;
results{loc}.name = 'EM-BiG-AMP (Rank Contraction)';
results{loc}.err = opt.error_function(estFin.Ahat*estFin.xhat);
results{loc}.time = tGAMP;
results{loc}.errHist = estHistEM.errZ;
results{loc}.timeHist = estHistEM.timing;
results{loc}.rank = size(estFin.xhat,1);
end
Starting EM-BiG-AMP with rank contraction
It 0001 nuX = 3.962e-01 meanX = 0.000e+00 tol = 1.000e-04 nuw = 9.906e-02 SNR = 20.00 Error = -16.0272 numIt = 0050
Updating rank estimate from 25 to 10 on iteration 1
It 0002 nuX = 3.458e-01 meanX = 0.000e+00 tol = 1.000e-04 nuw = 9.715e-02 SNR = 19.39 Error = -38.1882 numIt = 0045
It 0003 nuX = 5.983e-01 meanX = 0.000e+00 tol = 4.811e-05 nuw = 4.763e-04 SNR = 43.18 Error = -69.8033 numIt = 0030
It 0004 nuX = 6.026e-01 meanX = 0.000e+00 tol = 2.016e-08 nuw = 2.017e-07 SNR = 76.95 Error = -135.1314 numIt = 0053
It 0005 nuX = 6.026e-01 meanX = 0.000e+00 tol = 1.000e-08 nuw = 5.181e-14 SNR = 142.86 Error = -160.3864 numIt = 0030
It 0006 nuX = 6.026e-01 meanX = 0.000e+00 tol = 1.000e-08 nuw = 5.197e-14 SNR = 168.11 Error = -185.3936 numIt = 0030
Try Matrix ALPS
if tryMatrixAlps
disp('Starting Matrix ALPS')
Alpsparams.ALPSiters = 1500;
Alpsparams.tol = opt.tol;
Alpsparams.xpath = 0;
Alpsparams.svdMode = 'propack';
Alpsparams.cg_tol = 1e-10;
Alpsparams.cg_maxiter = 500;
Alpsparams.svdApprox = 0;
Alpsparams.power = 2;
Alpsparams.params.tau = 0;
[ix,iy] = find(omega);
tlinearInd = sub2ind([L M], double(iy), double(ix));
tidx = tlinearInd(:);
tA = @(z) z(tidx);
tAt = @(z) full(sparse(double(iy), double(ix), z, L, M));
yt = tA(Y');
tstart = tic;
[X_hat3, numiter3, X_path3,estHistMatrixAlps,timingMatrixAlps] =...
matrixALPSII_QR_timing(yt, tA, tAt, L, M, N, Alpsparams, [],error_function);
tMatrixAlps = toc(tstart);
loc = length(results) + 1;
results{loc}.name = 'Matrix ALPS';
results{loc}.err = estHistMatrixAlps.errZ(end);
results{loc}.time = tMatrixAlps;
results{loc}.errHist = estHistMatrixAlps.errZ;
results{loc}.timeHist = timingMatrixAlps;
end
Starting Matrix ALPS
Try Grouse
if tryGrouse
disp('Starting GROUSE')
step_size = 0.5;
maxCycles = 600;
maxrank = N;
[I,J] = find(omega);
S = reshape(Y(omega),[],1);
tstart = tic;
[~,~,~,timingGrouse,estHistGrouse] =...
grouse_timing(I,J,S,M,L,maxrank,step_size,maxCycles,opt.tol,error_function);
tGrouse = toc(tstart);
loc = length(results) + 1;
results{loc}.name = 'GROUSE';
results{loc}.err = estHistGrouse.errZ(end);
results{loc}.time = tGrouse;
results{loc}.errHist = estHistGrouse.errZ;
results{loc}.timeHist = timingGrouse;
end
Starting GROUSE
Try VBMC
if tryVbmc
options.verbose = 0;
options.MAXITER = 2000;
options.DIMRED = 0;
options.UPDATE_BETA = 0;
options.beta = 1e9;
options.initial_rank = N;
disp('Starting VBMC');
tstart = tic;
[~,timingVbmc,estHistVbmc] = VBMC_timing(omega, Y, options,error_function);
tVbmc = toc(tstart);
loc = length(results) + 1;
results{loc}.name = 'VSBL';
results{loc}.err = estHistVbmc.errZ(end);
results{loc}.time = tVbmc;
results{loc}.errHist = estHistVbmc.errZ;
results{loc}.timeHist = timingVbmc;
end
Starting VBMC
Try LMaFit
if tryLmafit
disp('Starting LMaFit')
Known = find(omega);
data = reshape(Y(omega),[],1);
Lmafit_opts.est_rank = 0;
Lmafit_opts.tol = opt.tol;
Lmafit_opts.print = 0;
Lmafit_opts.maxit = 6000;
Lmafit_opts.init = 0;
Lmafit_opts.X = Ahat;
Lmafit_opts.Y = xhat;
tstart = tic;
[~,~,~,timingLmafit,estHistLmafit] =...
lmafit_mc_adp_timing(M,L,N,Known,data,Lmafit_opts,error_function);
tLmafit = toc(tstart);
loc = length(results) + 1;
results{loc}.name = 'LMaFit';
results{loc}.err = estHistLmafit.errZ(end);
results{loc}.time = tLmafit;
results{loc}.errHist = estHistLmafit.errZ;
results{loc}.timeHist = timingLmafit;
end
Starting LMaFit
Try inexact ALM
if tryInexactAlm
disp('Starting Inexact ALM')
try
tstart = tic;
[~,~,~,timingInexactAlm,estHistInexactAlm] =...
inexact_alm_mc_tasos_timing(sparse(Y),opt.tol,2000,N,error_function);
tInexactAlm = toc(tstart);
loc = length(results) + 1;
results{loc}.name = 'Inexact ALM';
results{loc}.err = estHistInexactAlm.errZ(end);
results{loc}.time = tInexactAlm;
results{loc}.errHist = estHistInexactAlm.errZ;
results{loc}.timeHist = timingInexactAlm;
catch
loc = length(results) + 1;
results{loc}.name = 'Inexact ALM';
results{loc}.err = inf;
results{loc}.time = inf;
results{loc}.errHist = inf;
results{loc}.timeHist = inf;
end
end
Starting Inexact ALM
Store the options structures in results
results{1}.optIn = optIn;
Show Results
if nargin == 0
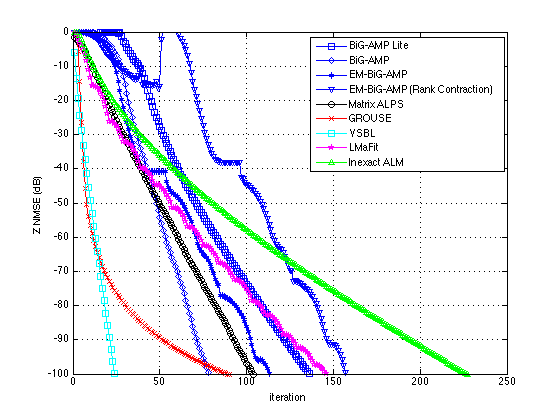
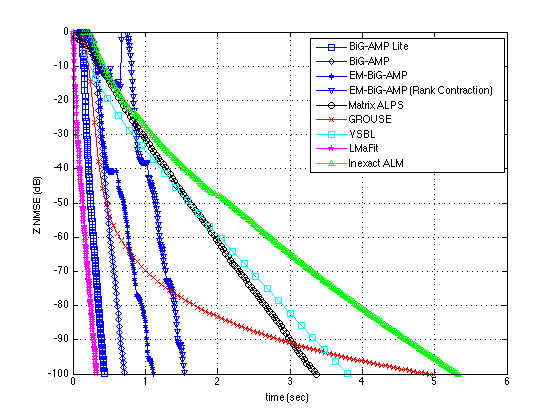
plotUtilityNew(results,[-100 0],200,201)
results{:}
end
ans =
name: 'BiG-AMP Lite'
err: -127.8798
time: 1.6713
errHist: [176x1 double]
timeHist: [176x1 double]
optIn: [1x1 struct]
ans =
name: 'BiG-AMP'
err: -143.9354
time: 1.6218
errHist: [105x1 double]
timeHist: [105x1 double]
ans =
name: 'EM-BiG-AMP'
err: -152.8152
time: 2.6799
errHist: [168x1 double]
timeHist: [168x1 double]
ans =
name: 'EM-BiG-AMP (Rank Contraction)'
err: -185.3936
time: 3.7089
errHist: [238x1 double]
timeHist: [238x1 double]
rank: 10
ans =
name: 'Matrix ALPS'
err: -148.1998
time: 6.5163
errHist: [155x1 double]
timeHist: [155x1 double]
ans =
name: 'GROUSE'
err: -118.5300
time: 22.2990
errHist: [247x1 double]
timeHist: [247x1 double]
ans =
name: 'VSBL'
err: -134.5088
time: 5.7639
errHist: [34x1 double]
timeHist: [34x1 double]
ans =
name: 'LMaFit'
err: -149.7031
time: 2.0092
errHist: [247x1 double]
timeHist: [247x1 double]
ans =
name: 'Inexact ALM'
err: -131.2548
time: 11.0982
errHist: [328x1 double]
timeHist: [328x1 double]
ans =
Columns 1 through 4
[1x1 struct] [1x1 struct] [1x1 struct] [1x1 struct]
Columns 5 through 8
[1x1 struct] [1x1 struct] [1x1 struct] [1x1 struct]
Column 9
[1x1 struct]